Mosaic Fertilizers opens vacancies for the Palmeirante (TO) Mixing Unit
In total, there are 18 job openings scheduled to begin in June this year; The project is still in progress
Researchers from 16 countries, including Brazil, carried out the sequencing of the reference genome of the Arabica coffee species, the most consumed throughout the world. Three researchers are from Embrapa Café and another eight from institutions that make up the Café Research Consortium, of which Embrapa is the coordinator. Scientific article published on the 15th in the scientific journal Nature Genetics presents unprecedented information regarding the genome and population genomics of this species, which reveals the history of diversification of currently planted cultivars.
Researcher Alan Andrade, from Embrapa Café, explains that the group of scientists, of which he is part, carried out a complete structural genetic mapping of the Coffea arabica, with the highest quality achieved to date. “With this we arrived at what we call a reference genome. In 2004, we were pioneers here in Brazil in the functional sequencing of the Arabica species genome. Now, with the structural, we begin to know the order of the genes within the DNA sequences and the intergenic regions that make up the genome, which is not possible to see in functional sequencing.” This made it easier to identify genes that give plants specific characteristics such as resistance to diseases and drought or coffee cherry size, as well as aroma and flavor.
Researcher Luiz Filipe Pereira, also from Embrapa Café, adds that important advances are already being achieved based on the results obtained. “As we have been immersed in this work for years, we are developing several studies focusing on Brazilian coffee farming using data from this study.”
He explained that the detailed genome makes it possible to identify genetic variations in DNA bases associated with phenotypic characteristics, such as disease resistance. “In this way, through the analysis of the plants’ DNA we are able to quickly select those that are resistant, accelerating improvement”, detailed Pereira.
Data from the new sequencing is also being applied to the development of technologies for coffee certification and traceability. The Study also included the participation of Embrapa Café researcher Lilian Padilha, who worked together with the Agronomic Institute (IAC) team.
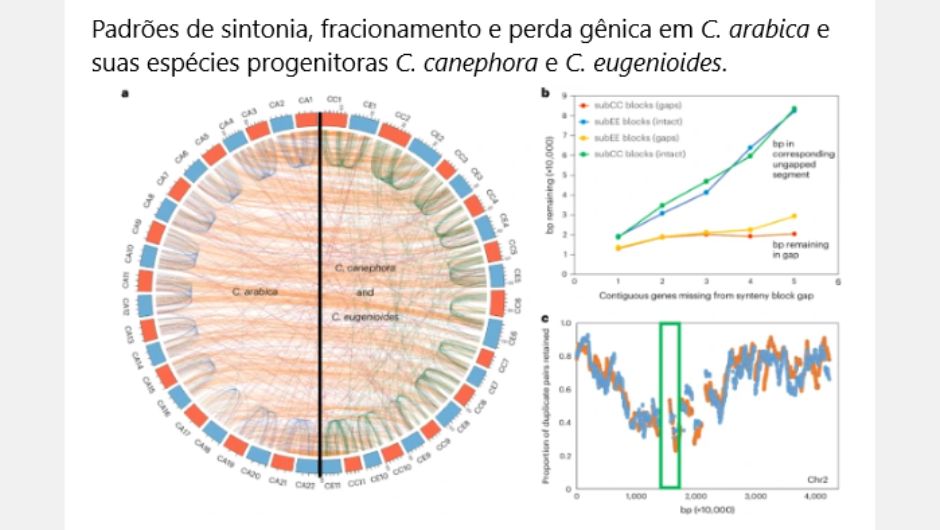
With the new genetic mapping, comparisons of the complete genome sequences and structures of the species were carried out Coffea arabica, Coffea eugenioides and again from Coffee Canephora. The objective was to reveal the evolution of species, the function of genes, the mechanism of gene regulation, identifying the sequence structures and elements that were conserved or differentiated. Analyzes were also carried out on the gene family, evolutionary development, duplication of the entire genome, and the selective pressure suffered.
According to the researchers, “modern genomic tools and a detailed understanding of the origin and breeding history of contemporary varieties are vital for the development of new Arabica coffee cultivars, better adapted to climate change and agricultural practices.”
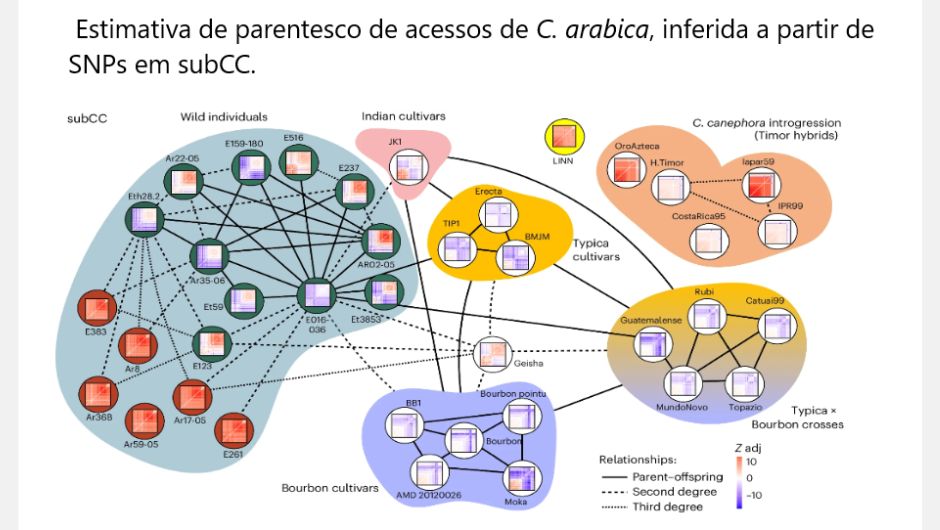
They re-sequenced the complete genome of 41 wild and cultivated accessions of this species, including an XNUMXth-century specimen used by Swedish naturalist Carl Linnaeus, which allowed an in-depth analysis of the history and routes of spread of the species. C. arabica.
A C. arabica It is a polyploid species, called allotetraploid, as it carries 44 chromosomes. It is the result of a natural hybridization event between the ancestors of the current Coffea canephora (Robusta coffee) and Coffea eugenioides, which have 22 chromosomes each, classified as diploid. This duplication of the entire genome is given the acronym WGD in English. Scientists had difficulty pinpointing exactly when – and where – this allopolyploidization event occurred, with estimates ranging between 10.000 and 1 million years ago.
Using computational modeling, the researchers sought signatures of the species' foundation by carrying out analyzes on the genomes of the species. C. arabica. The models show three population bottlenecks throughout history, the oldest of which occurred around 29 thousand generations ago, or 610 thousand years.
This suggests that Arabica was formed sometime between 360 and 610 years ago and had its population wax and wane in periods of warming and cooling of the Earth for thousands of years, before it was eventually cultivated in Ethiopia. and Yemen, and then spread across the globe.
It was previously believed that coffee plants were first cultivated in Ethiopia, but the varieties collected by researchers around the Great Rift Valley, which stretches from Southeast Africa to Asia, showed a clear geographic divide. The wild varieties studied all originate from the western side, while the cultivated varieties all originate from the eastern side, closer to the Bab al-Mandab Strait, which separates Africa from Yemen.
This would be in line with evidence that coffee cultivation may have started mainly in Yemen, around the 1600th century, and then moved to India, which supports the legend of the smuggling of “seven seeds” carried out by the Indian monk Baba Budan, around XNUMX. Thus, the diversity of Yemeni coffee may be the founder of all the main Arabica varieties today.
For scholars, polyploidy is a powerful evolutionary force that has shaped genome evolution in many eukaryotic lineages, possibly offering adaptive advantages in times of global change. However, contemporary Arabica cultivars descend from Typica or Bourbon strains, which have particularly low genetic diversity, are susceptible to many pests and diseases, such as coffee rust, and can be cultivated successfully only in some regions of the world.
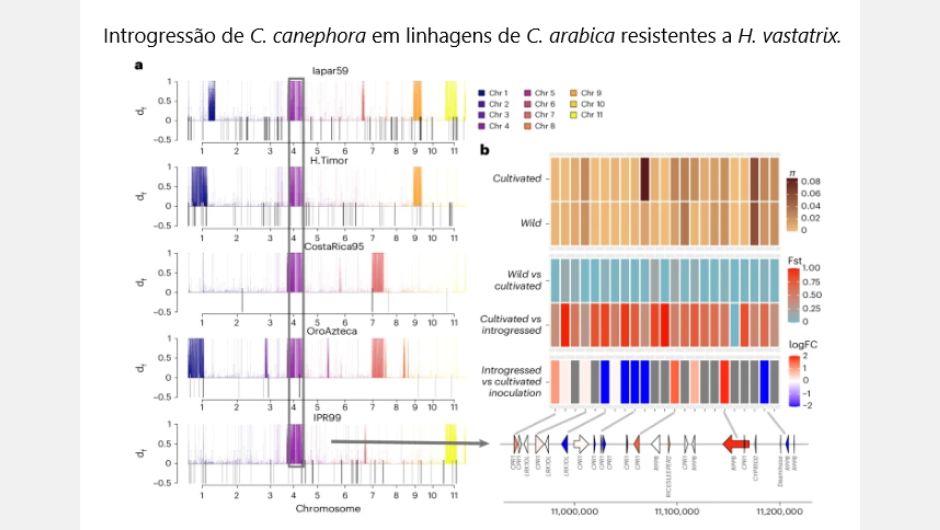
In 1927, a spontaneous hybrid of C. canephora was identified on the island of Timor, resistant to the fungus H. uvatrix, which causes rust. Using the new Arabica reference genome, studies carried out with plants of this lineage made it possible to identify a new target site to potentially improve resistance to pathogens, such as this fungus. The new genome sequencing has provided other new discoveries, such as which wild varieties are closest to today's cultivated Arabica coffee. Scientists also discovered that the Typica variety, an old Dutch cultivar originating from India or Sri Lanka, is probably the mother of the Bourbon variety, widely used in the preparation of specialty coffees.
Since the beginning of the 300th century, Brazil has led the world in the production and export of coffee, which has been present in the country for almost 1923 years. This leadership has been anchored by extensive research work linked to coffee farming, which dates back to the creation of the Coffee Section at the IAC in XNUMX. Since then, the country has continued to carry out studies linked to this culture.
A few years later, in 1929, with the creation of the Genetics Section, work on coffee genetics and improvement began. Since then, dozens of institutions have started to carry out studies for the coffee sector or were created based on it, such as Embrapa Café and the Consórcio Pesquisa Café, which currently brings together around 40 research bodies that carry out work focused on this crop.
In relation to the genetic sequencing of the coffee plant, Embrapa has made important advances. In 2004, Alan Andrade, Carlos Colombo, a researcher at IAC, and Luiz Gonzaga, a researcher at the Institute of Rural Development of Paraná (Iapar), coordinated the work on the first functional sequencing of the Arabica coffee genome, in a project by the Café Research Consortium, supported by the São Paulo State Research Support Foundation (Fapesp), which also included the participation of Luiz Filipe Pereira, and which generated at the time the largest database for coffee in the world, with 200 thousand DNA sequences.
The result of this work was decisive for the first total sequencing of the Coffea canephora, in work carried out by an international consortium made up of 11 countries, with significant participation from Andrade and Pereira.
Another important sequence was that of the genome of the leafminer, one of the main pests of the coffee plant, completed in 2022 in a project led by researchers from Embrapa Genetic Resources and Biotechnology, with the participation of researchers from Embrapa Agroindústria Tropical (CE), Embrapa Café (DF ), Embrapa Cerrados (DF), Embrapa Milho e Sorgo (MG) and Federal University of Viçosa (UFV).

Receive the latest agriculture news by email
In total, there are 18 job openings scheduled to begin in June this year; The project is still in progress
With more than 420 thousand properties regularized in 2023, the State Government is campaigning to speed up the verification of another 260 thousand pending registrations